Real Example Analysis
Overview
Teaching: 10 min
Exercises: 10 minQuestions
How do you visualize data from a
DataFrame?How do you group data by year and month?
How do you plot multiple measurements in a single plot?
Objectives
Learn how to plot the cleaned data
Learn how to subset and plot the data
Learn how to using the groupby method to visualize yearly and monthly changes
Analyzing Real Data
This episode continues from the previous one and utilizes the final DataFrame described there.
Analysis
Before we get started on our analysis let us take stock of how much data we have for the various columns. To do this we can use two DataFrame methods that we’ve previously used.
First we can check how many rows of data we have in total. We can check this easily through the shape attribute of the DataFrame
df.shape
(21222, 13)
From this we can see that we have 21222 rows in our data and 13 columns. So at most we can have 21222 rows of data for each column. However, as we saw during the cleaning phase there are NaN values in our dataset so many of our columns won’t contain data in every row.
To check how many rows of data we have for each column we can again use the describe() method. It will count how many row of data are not NaN for each column. To reduce the size of the output we will use loc to only view the counts for each column.
df.describe().loc["count",:]
time hhmmss 21222.0
press dbar 21222.0
temp ITS-90 21222.0
csal PSS-78 21210.0
coxy umol/kg 3727.0
ph 885.0
phos umol/kg 2259.0
nit umol/kg 2253.0
doc umol/kg 868.0
hbact #*1e5/ml 750.0
pbact #*1e5/ml 749.0
sbact #*1e5/ml 750.0
Name: count, dtype: float64
As we can see we have highly variable amounts of data for each of our column. We will ignore pressure since it is roughly a depth estimate. These fit fairly neatly into three groups:
- Data that is found in almost all rows
- Temperature
- Salinity
- Data that is found in around 2000-4000 samples
- Oxygen
- Phosphorus
- Nitrate+Nitrite
- Data that is found in fewer than 1000 samples
- pH
- Dissolved organic carbon
- Heterotrophic bacteria
- Prochlorococcus
- Synechococcus
Note here that pressure is roughly akin to “depth” so we won’t be using it.
GroupBy and Visualization
To start off we can focus on the measurements that we have plenty of data for.
Plotting Temperature
We can quickly get a matplotlib visualization for temperature by calling the
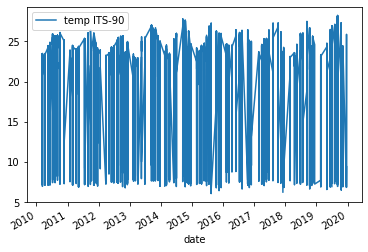
plotmethod from ourDataFrame. We can tell it what columns we want to use as the x axis and y axis via the parametersxandy. Thekindparameter lets theplot()method know what kind of plot we want e.g. line or scatter. For more information about the plot function check out the docs (Link to plot method docs).df.plot(x="date", y="temp ITS-90", kind="line")Solution
Below is the output plot
However, the plot we get is very messy. We see a lot of variation from around 25°C to 7°C year to year and the lines are clustered very tightly together.
Plotting Surface Temperature
Part of the reason we got this messy plot is because we are utilizing all the temperature values in our dataset, regardless of depth (i.e. pressure). To resolve some of the variation we can ask Pandas to only plot data that is from roughly the top 100m of the water column this would be roughly any rows that come from pressures of less than 100 dbar.
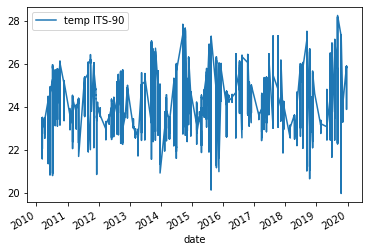
surface_samples = df[df["press dbar"] < 100] surface_samples.plot(x="date", y="temp ITS-90", kind="line")Solution
Below is the output plot
Now we can see that we have removed some of the variation we saw in the previous figure. However, it is still somewhat difficult to make out any trends in the data. One way of dealing with this would be to e.g. get the average temperature for each year and then plot those results.
To this we will introduce a new method called groupby which allows us to run calculations like mean() on groups we specify. For us we want to get the mean temperature for each year. Thanks to our previous work in setting up the date column type this is very easy. We can also reuse surface_samples to only get samples from the upper 100m of the water column.
grouped_surface_samples = surface_samples.groupby(df.date.dt.year).mean()
time hhmmss press dbar temp ITS-90 csal PSS-78 coxy umol/kg \
date
2010 111193.170920 37.517015 24.438342 35.282562 213.306024
2011 108457.713156 40.025449 24.545986 35.225530 210.137566
2012 103491.885435 37.503730 24.286195 35.200273 211.909375
2013 110479.360743 38.544562 24.449899 35.302473 211.057143
2014 107638.631016 40.763369 24.755485 35.287073 211.538974
2015 108635.305970 37.930929 24.799153 35.210472 210.160287
2016 101880.099391 37.814300 24.940249 34.995771 208.771186
2017 106559.737030 36.499374 24.836940 35.000984 211.143750
2018 103767.267879 37.064727 24.580730 34.999648 213.029371
2019 104902.595131 38.461587 25.087561 34.805649 212.239130
ph phos umol/kg nit umol/kg doc umol/kg hbact #*1e5/ml \
date
2010 8.063846 0.075842 0.033465 73.810154 4.614946
2011 8.072089 0.056455 0.029464 74.384000 4.651204
2012 8.061535 0.114953 0.018224 72.578871 4.787081
2013 8.064927 0.087264 0.026981 71.548413 4.778000
2014 8.069786 0.065983 0.025652 72.237091 4.845439
2015 8.065829 0.071795 0.029211 70.894154 4.838659
2016 8.072237 0.068108 0.055676 72.562131 4.467324
2017 8.064918 0.052946 0.033514 70.556863 4.744976
2018 8.054656 0.060000 0.085250 NaN 4.555553
2019 8.061786 0.060093 0.021667 NaN 5.084833
pbact #*1e5/ml sbact #*1e5/ml
date
2010 1.909351 0.014351
2011 1.783857 0.019388
2012 1.968270 0.015351
2013 2.345216 0.020162
2014 2.223146 0.018927
2015 2.182341 0.018386
2016 2.127529 0.014471
2017 2.109878 0.019927
2018 2.035622 0.020711
2019 2.123119 0.015214
We see now that the new DataFrame generated by groupby() and mean() contains the mean for each year for each of our columns.
Plotting Yearly Surface Temperature
Now we can just run the same plot method as previously but using
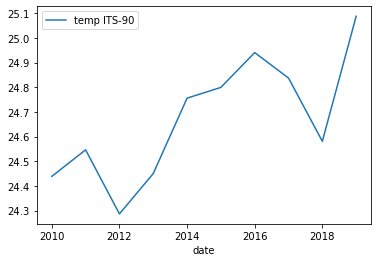
grouped_surface_samplesinstead ofsurface_samples.grouped_surface_samples.plot(x="date", y="temp ITS-90", kind="line")Solution
Below is the output plot
Now it looks a lot smoother, but now we have another issue. We’ve smoothed out any month to month variations that are present in the data. To fix this we can instead use the groupby method to group by year and month.
grouped_surface_samples = surface_samples.groupby([(surface_samples.date.dt.year),(surface_samples.date.dt.month)]).mean()
Plotting Monthly Surface Temperature
If we plot this we get a month by month plot of temperature variations.
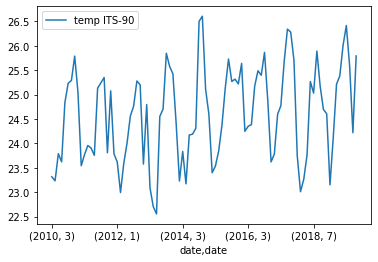
grouped_surface_samples.plot(y="temp ITS-90", kind="line")Solution
Below is the output plot
While we have been focusing on temperature there is no reason that we can’t redo the same plots that we have been making with measurements other than temperature. We can also plot multiple measurements at the same time if we want to as well.
Plotting Monthly Surface Temperature
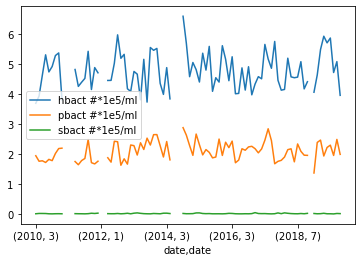
To test this we will try plotting the abundance of Prochlorococcus, Synechococcus, and heterotrophic bacteria.
grouped_surface_samples.plot(y=["hbact #*1e5/ml", "pbact #*1e5/ml", "sbact #*1e5/ml"], kind="line")Solution
Below is the output plot
With that we plotted looked various methods of plotting the data we have in our dataset. We’ve also learned how to group different measurements depending on when the measurement was taken. If you are interested you can keep testing different methods of grouping the data or plotting some of the measurements that we did not use e.g. pH or dissolved organic carbon (doc umol/kg).
Hopefully throughout this lesson you have learned some useful skills in order to both analyze your data and document your analysis and any code that you used. There is plenty of things that we did not have time to go over so make sure to keep learning!
Key Points
Grouping data by year and months is a powerful way to identify monthly and yearly changes
You can easily add more measurements to a single plot by using a list
There is a lot we didn’t cover here, so take a look at the Matplotlib docs (Link to Matplotlib docs) and other libraries that can allow you to make dynamic plots e.g. Plotly (Link to Plotly docs)