Introduction
Overview
Teaching: 8 min
Exercises: 0 minQuestions
Who are we and what are we going to learn?
Objectives
Introduce ourselves and the course
Setup Hydroshare our example FAIR data platform
Better research by better sharing
Introductions
Introductions
- Sean Cleveland, Cyberinfrastructure Scientist, The University of Hawai’i Cyberinfrastructure & Hawaii Data Science Institute
- Bjarne Bartlett, CI-TRACS Data Science Fellow, The University of Hawai’i Cyberinfrastructure & Hawaii Data Science Institute
Hello everyone, and welcome to the FAIR Data Management Security and Ethics workshop.
Better research by better sharing
For many of us, data management or output sharing in general are considered a burden rather than a useful activity. Part of the problem is our bad timing and lack of planning.
Data management is a continuous process
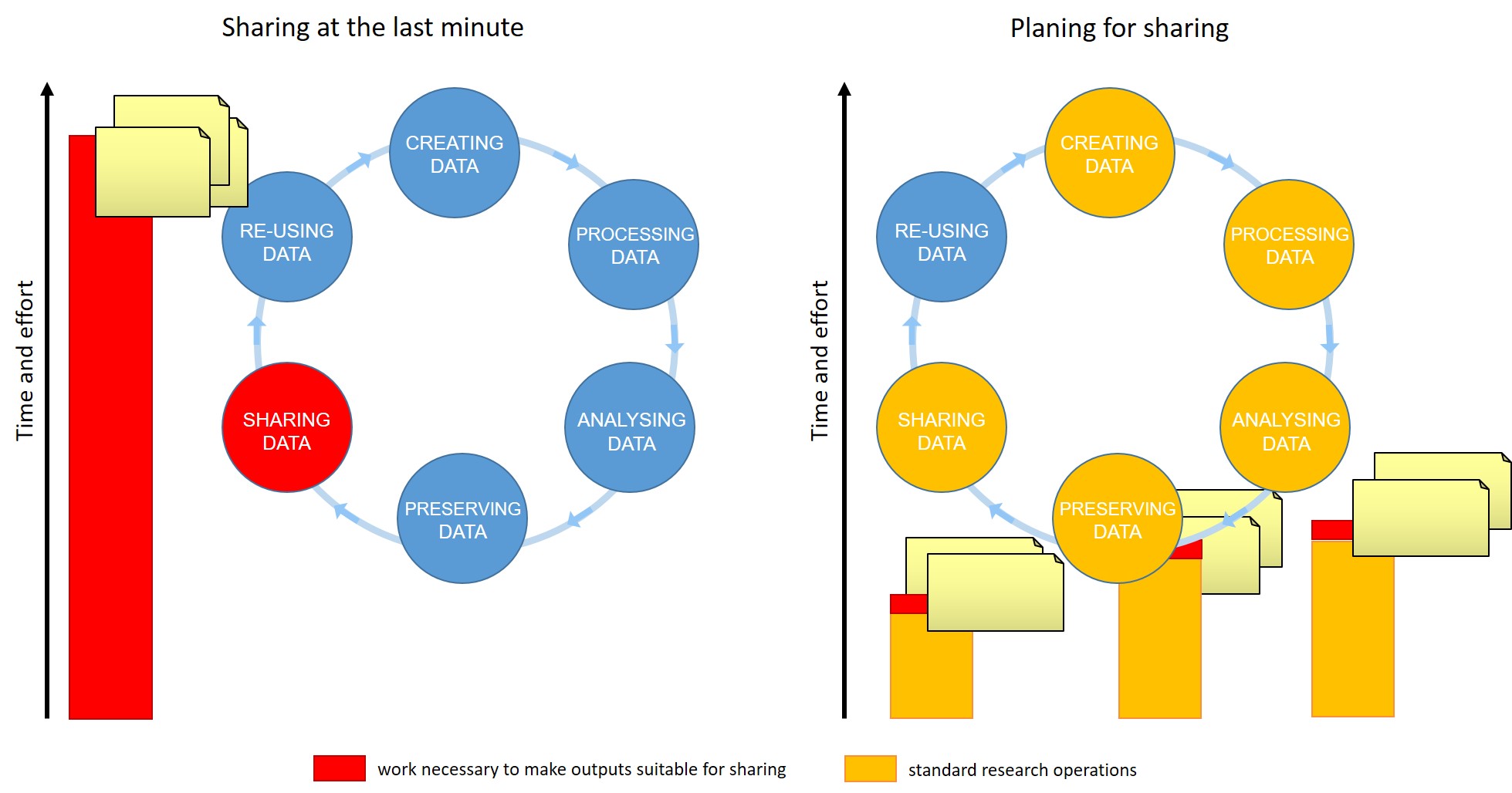 Figure credits: Tomasz Zielinski and Andrés Romanowski from https://carpentries-incubator.github.io/fair-bio-practice/06-being-precise/index.html
Figure credits: Tomasz Zielinski and Andrés Romanowski from https://carpentries-incubator.github.io/fair-bio-practice/06-being-precise/index.html
When should you engage in data sharing and open practices?
- Data management should be done throughout the duration of your project.
- If you wait till the end, it will take a massive effort on your side and will be more of a burden than a benefit.
- Taking the time to do effective data management will help you understand your data better and make it easier to find when you need it (for example when you need to write a manuscript or a thesis!).
- All the practices that enable others to access and use your outcomes directly benefit you and your group
In this workshop we will discuss how your research outputs can be made readily available for re-use by others.
Key Points
You can do more impactful research if you plan to share your outputs!
You can more efficiently publish if you plan to share your outputs!
Open Science
Overview
Teaching: 8 min
Exercises: 4 minQuestions
What is Open Science?
How can I benefit from Open Science?
Why has Open Science become a hot topic?
Objectives
Identify parts of the Open Science movement, their goals and motivations
Explain the main benefits of Open Science
Recognize the barriers and risks in the adoption of Open Science practices
Science works best by exchanging ideas and building on them. Most efficient science involves both questions and experiments being made as fully informed as possible, which requires the free exchange of data and information.
All practices that make knowledge and data freely available fall under the umbrella-term of Open Science/Open Research. It makes science more reproducible, transparent, and accessible. As science becomes more open, the way we conduct and communicate science changes continuously.
What is Open Science
Open science is the movement to make scientific research (including publications, data, physical samples, and software) and its dissemination accessible to all levels of an inquiring society, amateur or professional.
Open Science represents a new approach to the scientific process based on cooperative work and new ways of diffusing knowledge by using digital technologies and new collaborative tools
Open science is transparent and accessible knowledge that is shared and developed through collaborative networks.
Characteristics:
- Using web-based tools to facilitate information exchange and scientific collaboration
- Transparency in experimental methodology, observation, and collection of data
- Public availability and reusability of scientific data, methods and communications
What is the Open Science movement?
Sharing of information is fundamental for science. This began at a significant scale with the invention of scientific journals in 1665. At that time this was the best available alternative to critique & disseminate research, and foster communities of like-minded researchers.
Whilst this was a great step forward, the journal-driven system of science has led to a culture of ‘closed’ science, where knowledge or data is unavailable or unaffordable to many.
The distribution of knowledge has always been subject to improvement. Whilst the internet was initially developed for military purposes, it was hijacked for communication between scientists, which provided a viable route to change the dissemination of science.
The momentum has built up with a change in the way science is communicated to reflect what research communities are calling for – solutions to the majority of problems (e.g. impact factors, data reusability, reproducibility crisis, trust in the public science sector etc…) that we face today.
Open Science is the movement to increase transparency and reproducibility of research, through using the open best practices.
Attribution Gema Bueno de la Fuente
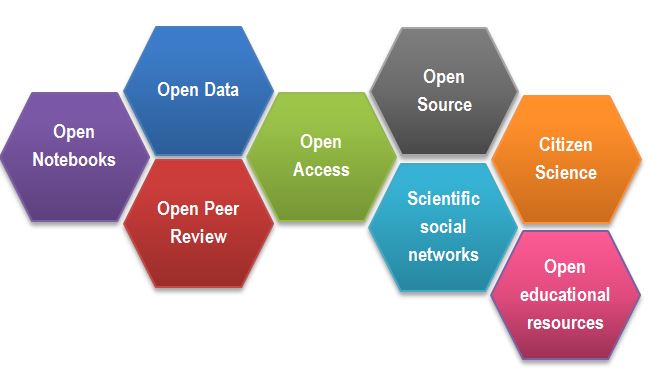
Open Science Building Blocks
-
Open Access: Research outputs hosted in a way that make them accessible for everyone. Traditionally Open Access referred to journal articles, but now includes books, chapters or images.
-
Open Data: Data freely and readily available to access, reuse, and share. Smaller data sets were often accessible as supplemental materials by journals alongside articles themselves. However, they should be hosted in dedicated platforms for more convenient and better access.
-
Open Software: Software where the source code is made readily available; others are free to use, change, and share. Some examples of these including the coding language and supporting software R and RStudio, as well as image analysis software such as Fiji/ImageJ.
-
Open Notebooks: Lab & notebooks hosted online, readily accessible to all. These are popular among some of the large funding bodies and allow anyone to comment on any stage of the experimental record.
-
Open Peer Review: A system where peer review reports are published alongside the body of work. This can include reviewers’ reports, correspondence between parties involved, rebuttals, editorial decisions etc…
-
Citizens Science: Lay people become involved in scientific research, most commonly in data collection or image analysis. Platforms such as zooniverse.org help connect projects with lay people interested in playing an active role in research, which can help generate and/or process data which would otherwise be unachievable by one single person.
-
Scientific social networks: Networks of researchers, which often meet locally in teams, but are also connected online, foster open discussions on scientific issues. Online, many people commonly use traditional social media platforms for this, such as Twitter, Instagram, various sub-reddits, discussion channels on Slack/Discord etc…, although there are also more dedicated spaces such as researchgate.net.
-
Open Education resources: Educational materials that are free for anyone to access and use to learn from. These can be anything from talks, instructional videos, and explanations posted on video hosting websites (e.g. YouTube), to entire digital textbooks written and then published freely online.
-
Citizen science: Citizen participation of various stages of research process from project funding to collecting and analysing data.
Exercise 1: Benefits of Open Science
Being open has other outcomes/consequences beyond giving the free access to information. For example, Open educational resources:
- enables collaborative development of courses
- improves teachers/instructors skills by sharing ideas
Select one or two of the following OS parts:
- Open Access
- Open Data
- Open Software
- Open Notebooks
- Open Peer Review
and discuss what are the benefits or what problems are solved by adaption of those Open initiatives.
Solution
Possible benefits and consequences for each part:
Open Access
- speed of knowledge distribution
- leveling field for underfunded sites which otherwise wouldn’t be able to navigate the paywall
- prevent articles being paid for ‘thrice’ (first to produce, second to publish, third to access) by institutions.
- greater access to work by others, increasing chance for exposure & citations
- access to work by lay audiences, thus increases social exposure of research
Open Data
- ensures data isn’t lost overtime - reusability
- acceleration of scientific discovery rate
- value for money/reduced redundancy
- permits statistical re-analysis of the data to validate findings
- gives access to datasets which were not published as papers (e.g. negative results, large screening data sets)
- provides an avenue to generate new hypotheses
- permits combination of multiple data sources to address questions, provides greater power than a single data source
Open Software
- great source to learn programming skills
- the ability to modify creates a supportive community of users and rapid innovation
- saves time
- faster bug fixes
- better error scrutiny
- use of the same software/code allows better reproducibility between experiments
- need funds to maintain and update software
Open Notebooks
- 100% transparent science, allowing input from others at early stages of experiments
- source of learning about the process of how science is actually conducted
- allows access to experiments and data which otherwise never get published
- provides access to ‘negative’ results and failed experiments
- anyone, anywhere around the world, at any time, can check in on projects, including many users simultaneously
- possibility of immediate feedback
- thorough evidence of originality of ideas and experiments, negating effect of ‘scooping’
Open Peer Review
- visibility leads to more constructive reviews
- mitigates against editorial conflicts of interest and/or biases
- mitigates against reviewers conflicts of interest and/or biases
- allows readers to learn/benefit from comments of the reviewers
Open Educational Materials
- Foster collaboration between educators/others
- Show clearly how method was taught (e.g. Carpentries materials) which can be reproduces anywhere, anytime
- protects materials from becoming technologically obsolete
- authors preparing the material or contribute all earn credit (e.g. GitHub)
- recycle animations and material that is excellent (why reinvent the wheel?)
Motivation: Money
One has to consider the moral objectives that accompany the research/publication process: charities/taxpayers pay to fund research, these then pay again to access the research they already funded.
From an economic point of view, scientific outputs generated by public research are a public good that everyone should be able to use at no cost.
According to EU report “Cost-benefit analysis for FAIR research data”, €10.2bn is lost every year because of not accessible data (plus additional 16bn if accounting for re-use and research quality).
The goals of Open Science is to make research and research data available to e.g. charities/taxpayers who funded this research.
Motivation: Reproducibility
The inherited transparency of Open Science and the easy access to data, methods and analysis details naturally help to address part of the Reproducibility crisis. The openness of scientific communications and of the actual process of evaluation of the research (Open Peer Review) increases confidence in the research findings.
Personal motivators
Open Science is advantageous to many parties involved in science (including researcher community, funding bodies, the public even journals), which is leading to a push for the widespread adoption of Open Science practices.
Funding bodies are also becoming big supporters of Open Science. We can see with the example of Open Access, that once enforced by funders (the stick) there is a wide adoption. But what about the personal motivators, the carrots.
The main difference between the public benefits of Open Science practices and the personal motivators of outputs creators, that the public can benefit almost instantly from the open resources. However, the advantages for data creator comes with a delay, typically counted in years. For example, building reputation will not happen with one dataset, the re-use also will lead to citations/collaboration after the next research cycle.
Barriers and risks of OS movement:
Exercise 2: Why we are not doing Open Science already
Discuss Open Science barriers, mention the reasons for not already being open:
Solution
- sensitive data (anonymising data from administrative health records can be difficult)
- IP
- misuse (fake news)
- lack of confidence (the fear of critics)
- lack of expertise
- the costs in $ and in time
- novelty of data
- it is not mandatory
- lack of credit (publishing negative results is of little benefit to you)
It may seem obvious that we should adopt open science practices, but there are associated challenges with doing so.
Sensitivity of data is sometimes considered a barrier. Shared data needs to be compliant with data privacy laws, leading many to shy away from hosting it publicly. Anonymising data to desensitise it can help overcome this barrier.
The potential for intellectual property on research can dissuade some from adopting open practices. Again, much can be shared if the data is filtered carefully to protect anything relating to intellectual property.
Another risk could be seen with work on Covid19: pre-prints. A manuscript hosted publicly prior to peer review, may accelerate access to knowledge, but can also be misused and/or misunderstood. This can result in political and health decision making based on faulty data, which is counter to societies’ best interest.
One concern is that opening up ones data to the scientific community can lead to the identification of errors, which may lead to feelings of embarrassment. However, this could be considered an upside - we should seek for our work to be scrutinized and errors to be pointed out, and is the sign of a competent scientist. One should rather have errors pointed out rather than risking that irreproducible data might cause even more embarrassment and disaster.
One of the biggest barriers are the costs involved in “being Open”. Firstly, making outputs readily available and usable to others takes time and significant effort. Secondly, there are costs of hosting and storage. For example, microscopy datasets reach sizes in terabytes, making such data accessible for 10 years involves serious financial commitment.
Where to next
Further reading/links:
Attribution
Content of this episode was adapted from:
- Wiki Open Science
- European Open Science Cloud
- Science is necessarily collaborative - The Biochemist article.
- @@(https://carpentries-incubator.github.io/fair-bio-practice/)
Key Points
Open Science increases transparency in research
Publicly funded science should be publicly available
Intellectual Property, Licensing, and Openness
Overview
Teaching: 8 min
Exercises: 2 minQuestions
What is intellectual property?
Why should I consider IP in Open Science?
Objectives
Timeline matters for legal protection
Understand what can and cannot be patented
Understand what licenses to use for re-use of data and software
Open Science and Intellectual property
Intellectual property (IP) is something that you create using your mind - for example, a story, an invention, an artistic work or a symbol.
The timeline of “opening” matters when one seeks legal protection for their IP.
For example, patents are granted only for inventions that are new and were not known to the public in any form. Publishing in a journal or presenting in a conference information related to the invention completely prevents the inventor from getting a patent!
You can benefit from new collaborations, industrial partnerships, and consultations which are acquired by openness. This can yield greater benefit than from patent-related royalties.
(Optional) Intellectual property protection
You can use a patent to protect a non-obvious (technical) invention that provides “technical contribution” or solves a “technical problem”. It gives you the right to take legal action against anyone who makes, uses, sells or imports it without your permission.
In principle, software can be patented. It is usually, settled by the court for each case.
Software code is copyrighted. Copyright prevents people from:
- copying your code
- distributing copies of it, whether free of charge or for sale.
Data cannot be patented, and in principle, it cannot be copyrighted. It is not possible to copyright facts!
Facts are not patentable, and since machine learning algorithms like neural networks are basically mathematical methods, they are exempt from protection. However, applied to a certain problem, an algorithm may become part of a patent. IF framed it in the right way, patenting an algorithm is possible. For example, a deep learning algorithm generating a certain kind of audio may be eligible. But that would not prevent the network from being applied to any other problem.
However, how data are collated and presented (especially if it is a database), can have a layer of copyright protection. Deciding what data needs to be included in a database, how to organize the data, and how to relate different data elements are all creative decisions that may receive copyright protection. Again, it is often a case by case situation and may come down to who has better lawyers.
After:
Exercise 3: Checking common licenses
Open CC BY license summary https://creativecommons.org/licenses/by/4.0/ is it clear how you can use the data under this licence and why it is popular in academia?
Check the MIT license wording: https://opensource.org/licenses/MIT is it clear what you can do with software code under this licence?
Compare the full wording of CC BY https://creativecommons.org/licenses/by/4.0/legalcode can you guess why the MIT licence is currently the most popular for open source code?
Solution
- CC BY license states material can be reproduced, shared, in whole or in part, unless where exceptions and limitations are stated. Attributions must be made to the Licensor.
- MIT license states that Software can by used without restriction (to copy, modify, publish, distribute etc…)
- The MIT license is short, to the point and optimised for software developers as it offers flexibility.
Attribution
Content of this episode was adapted from:
- @@(https://carpentries-incubator.github.io/fair-bio-practice/)
Key Points
A license is a promise not to sue - therefore attach license files
For data use Creative Commons Attribution (CC BY) license
For code use open source licenses such as MIT, BSD, or Apache license
FAIR Introduction
Overview
Teaching: 5 min
Exercises: 0 minQuestions
What are the FAIR principles?
Why should I care to be FAIR?
How do I get started?
Objectives
Identify the FAIR principles
Recognize the importance of moving towards FAIR in research
Relate the components of this lesson to the FAIR principles
What is FAIR?
The FAIR principles for research data, originally published in a 2016 Nature paper, are intended as “a guideline for those wishing to enhance the reusability of their data holdings.” This guideline has subsequently been endorsed by working groups, funding bodies and institutions.
FAIR is an acronym for Findable, Accessible, Interoperable, Reusable.
- Findable: others (both human and machines) can discover the data
- Accessible: others can access the data
- Interoperable: the data can easily be used by machines or in data analysis workflows.
- Re-usable: the data can easily be used by others for new research
The FAIR principles have a strong focus on “machine-actionability”. This means that the data should be easily readable by computers (and not only by humans). This is particularly relevant for working with and discovering new data.
What the FAIR principles are not
A standard: The FAIR principles need to be adopted and followed as much as possible by considering the research practices in your field.
All or nothing: making a dataset (more) FAIR can be done in small, incremental steps.
Open data: FAIR data does not necessarily mean openly available. For example, some data cannot be shared openly because of privacy considerations. As a rule of thumb, data should be “as open as possible, as closed as necessary.”
Tied to a particular technology or tool. There might be different tools that enable FAIR data within different disciplines or research workflows.
Why FAIR?
The original authors of the FAIR principles had a strong focus on enhancing reusability of data. This ambition is embedded in a broader view on knowledge creation and scientific exchange. If research data are easily discoverable and re-usable, this lowers the barriers to repeat, verify, and build upon previous work. The authors also state that this vision applies not just to data, but to all aspects of the research process.
What’s in it for you?
FAIR data sounds like a lot of work. Is it worth it? Here are some of the benefits:
- Funder requirements
- It makes your work more visible
- Increase the reproducibility of your work
- If others can use it easily, you will get cited more often
- You can create more impact if it’s easier for others to use your data
- …
Getting started with FAIR (climate) data
As mentioned above, the FAIR principles are intended as guidelines to increase the reusability of research data. However, how they are applied in practice depends very much on the domain and the specific use case at hand.
For the domain of climate sciences, some standards have already been developed that you can use right away. In fact, you might already be using some of them without realizing it. NetCDF files, for example, already implement some of the FAIR principles around data modeling. But sometimes you need to find your own way.
Challenge for yourself - Evaluate one of your own datasets
Pick one dataset that you’ve created or worked with recently, and answer the following questions:
- If somebody gets this dataset from you, would they be able to understand the structure and content without asking you?
- Do you know who has access to this dataset? Could somebody easily have access to this dataset? How?
- Does this dataset needs proprietary software to be used?
- Does this dataset have a persistent identifier or usage licence?
Attribution
Content of this episode was adapted from:
- @@(https://esciencecenter-digital-skills.github.io/Lesson-FAIR-Data-Climate/)
Key Points
The FAIR principles state that data should be Findable, Accessible, Interoperable, and Reusable.
FAIR data enhance impact, reuse, and transparancy of research.
FAIRification is an ongoing effort accross many different fields.
FAIR principles are a set of guiding principles, not rules or standards.
Findable
Overview
Teaching: 8 min
Exercises: 2 minQuestions
What is a persistent identifier or PID?
What types of PIDs are there?
Objectives
Explain what globally unique, persistent, resolvable identifiers are and how they make data and metadata findable
Articulate what metadata is and how metadata makes data findable
Articulate how metadata can be explicitly linked to data and vice versa
Understand how and where to find data discovery platforms
Articulate the role of data repositories in enabling findable data
For data & software to be findable:
F1. (meta)data are assigned a globally unique and eternally persistent identifier or PID
F2. data are described with rich metadata
F3. (meta)data are registered or indexed in a searchable resource
F4. metadata specify the data identifier
Persistent identifiers (PIDs) 101
A persistent identifier (PID) is a long-lasting reference to a (digital or physical) resource:
- Designed to provide access to information about a resource even if the resource it describes has moved location on the web
- Requires technical, governance and community to provide the persistence
- There are many different PIDs available for many different types of scholarly resources e.g. articles, data, samples, authors, grants, projects, conference papers and so much more
Different types of PIDs
PIDs have community support, organizational commitment and technical infrastructure to ensure persistence of identifiers. They often are created to respond to a community need. For instance, the International Standard Book Number or ISBN was created to assign unique numbers to books, is used by book publishers, and is managed by the International ISBN Agency. Another type of PID, the Open Researcher and Contributor ID or ORCID (iD) was created to help with author disambiguation by providing unique identifiers for authors. The ODIN Project identifies additional PIDs along with Wikipedia’s page on PIDs.
Digital Object Identifiers (DOIs)
The DOI is a common identifier used for academic, professional, and governmental information such as articles, datasets, reports, and other supplemental information. The International DOI Foundation (IDF) is the agency that oversees DOIs. CrossRef and Datacite are two prominent not-for-profit registries that provide services to create or mint DOIs. Both have membership models where their clients are able to mint DOIs distinguished by their prefix. For example, DataCite features a statistics page where you can see registrations by members.
Anatomy of a DOI
A DOI has three main parts:
- Proxy or DOI resolver service
- Prefix which is unique to the registrant or member
- Suffix, a unique identifier assigned locally by the registrant to an object
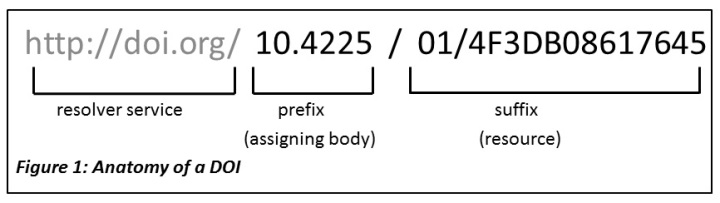
In the example above, the prefix is used by the Australian National Data Service (ANDS) now called the Australia Research Data Commons (ARDC) and the suffix is a unique identifier for an object at Griffith University. DataCite provides DOI display guidance so that they are easy to recognize and use, for both humans and machines.
Exercise 4
HydroShare is a data repository for water data from a variety of biological disciplines. It allows researchers to share and access water data for research. Visit the HydroShare resource search at Discover. Choose any dataset by clicking on the link. Now use control + F or command + F and search for ‘http’. Did the author use DOIs or persistent link from HydroShare for their data and software?
Solution
Authors will often link to platforms such as GitHub where they have shared their software and/or they will link to their website where they are hosting the data used in the paper. The danger here is that platforms like GitHub and personal websites are not permanent. Instead, authors can use repositories to deposit and preserve their data and software while minting a DOI. Links to software sharing platforms or personal websites might move but DOIs will always resolve to information about the software and/or data. See DataCite’s Best Practices for a Tombstone Page.
Rich Metadata
More and more services are using common schemas such as DataCite’s Metadata Schema or Dublin Core to foster greater use and discovery. A schema provides an overall structure for the metadata and describes core metadata properties. While DataCite’s Metadata Schema is more general, there are discipline specific schemas such as Data Documentation Initiative (DDI) and Darwin Core.
Thanks to schemas, the process of adding metadata has been standardized to some extent but there is still room for error. For instance, DataCite reports that links between papers and data are still very low. Publishers and authors are missing this opportunity.
Challenges: Automatic ORCID profile update when DOI is minted RelatedIdentifiers linking papers, data, software in Zenodo
Connecting research outputs
DOIs are everywhere. Examples.
Resource IDs (articles, data, software, …) Researcher IDs Organisation IDs, Funder IDs Projects IDs Instrument IDs Ship cruises IDs Physical sample IDs, DMP IDs… videos images 3D models grey literature
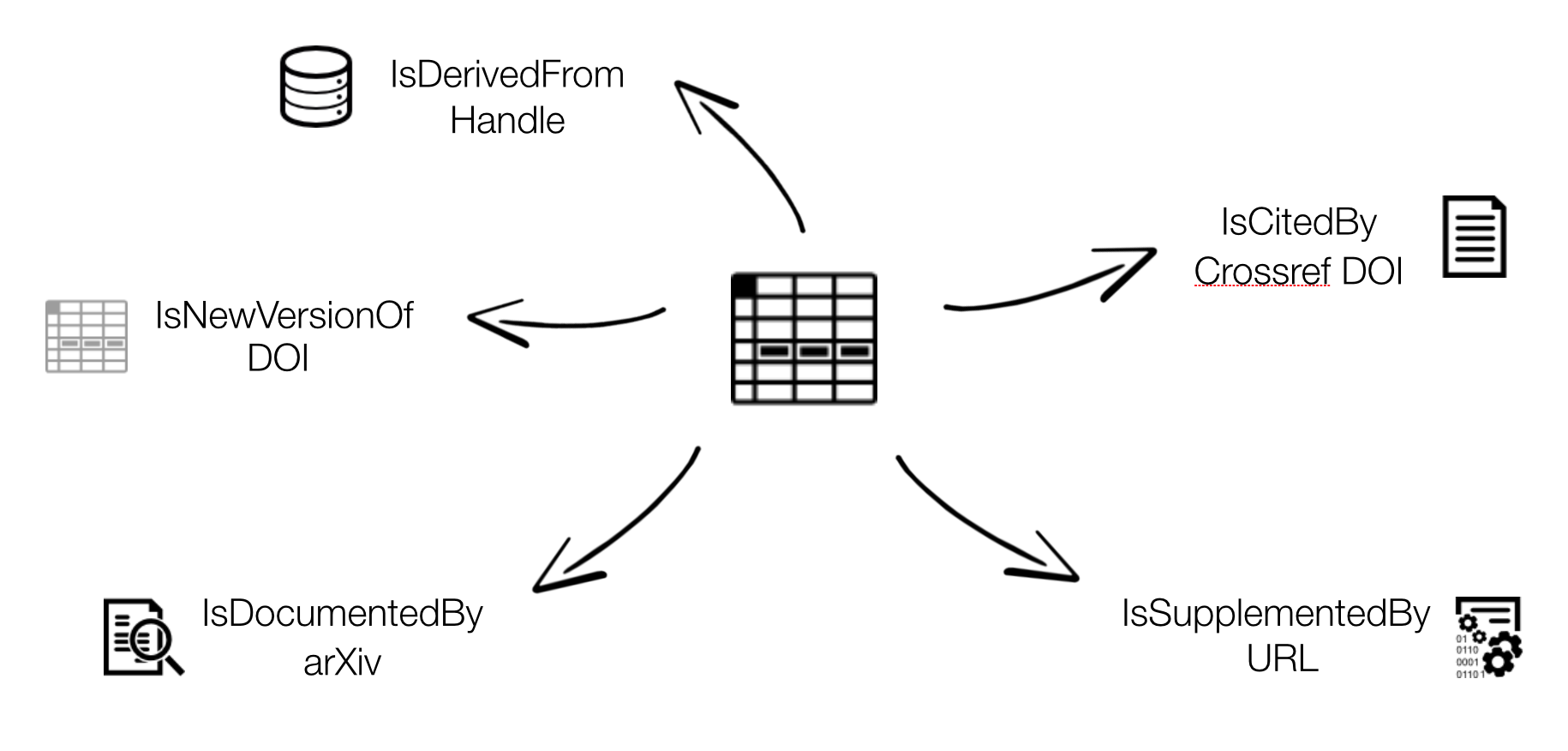
https://support.datacite.org/docs/connecting-research-outputs
Bullet points about the current state of linking… https://blog.datacite.org/citation-analysis-scholix-rda/
Provenance?
Provenance refers to the data lineage (inputs, entitites, systems, etc.) that ultimately impact validation & credibility. A researcher should comply to good scientific practices and be sure about what should get a PID (and what not). Metadata is central to visibility and citability – metadata behind a PID should be provided with consideration. Policies behind a PID system ensure persistence in the WWW - point. At least metadata will be available for a long time. Machine readability will be an essential part of future discoverability – resources should be checked and formats should be adjusted (as far possible). Metrics (e.g. altmetrics) are supported by PID systems.
Publishing behaviour of researchers
According to:
Technische Informationsbibliothek (TIB) (conducted by engage AG) (2017): Questionnaire and Dataset of the TIB Survey 2017 on information procurement and publishing behavior of researchers in the natural sciences and engineering. Technische Informationsbibliothek (TIB). DOI: https://doi.org/10.22000/54
- responses from 1400 scientists in the natural sciences & engineering (across Germany)
- 70% of the researchers are using DOIs for journal publications
- less than 10% use DOIs for research data – 56% answered that they don’t know about the option to use DOIs for other publications (datasets, conference papers etc.) – 57% stated no need for DOI counselling services – 40% of the questioned researchers need more information – 30% cannot see a benefit from a DOI
Choosing the right repository
Some things to check:
- Ask your colleagues & collaborators
- determining the right repo for your research
- That data are kept safe in a secure environment and data are regularly backed up and preserved (long-term) for future use
- Data can be easily discovered by search engines and included in online catalogues
- Intellectual property rights and licencing of data are managed
- Access to data can be administered and usage monitored
- That visibility of data can be enhanced to enable more use and citation
The decision for or against a specific repository depends on various criteria, e.g.
- Data quality
- Discipline
- Institutional requirements
- Reputation (researcher and/or repository)
- Visibility of research
- Legal terms and conditions
- Data value (FAIR Principles)
Some recommendations: → look for the usage of PIDs → look for the usage of standards (DataCite, Dublin Core, discipline-specific metadata → look for licences offered → look for certifications (DSA / Core Trust Seal, DINI/nestor, WDS, …)
Searching re3data w/ exercise https://www.re3data.org/ Out of more than 2115 repository systems listed in re3data.org in July 2018, only 809 (less than 39 %!) state to provide a PID service, with 524 of them using the DOI system
Search open access repos http://v2.sherpa.ac.uk/opendoar/
FAIRSharing https://fairsharing.org/databases/
Data Journals
Another method available to researchers to cite and give credit to research data is to author works in data journals or supplemental approaches used by publishers, societies, disciplines, and/or journals.
Articles in data journals allow authors to:
- Describe their research data (including information about process, qualities, etc)
- Explain how the data can be reused
- Improve discoverability (through citation/linking mechanisms and indexing)
- Provide information on data deposit
- Allow for further (peer) review and quality assurance
- Offer the opportunity for further recognition and awards
Examples:
- Nature Scientific data - published by Nature and established in 2013
- Geoscience Data Journal - published by Wiley and established in 2012
- Journal of Open Archaeology Data - published by Ubiquity and established in 2011
- Biodiversity Data Journal - published by Pensoft and established in 2013.
- Earth System Science Data - published by Copernicus Publications and established in 2009
Also, the following study discusses data journals in depth and reviews over 100 data journals: Candela, L. , Castelli, D. , Manghi, P. and Tani, A. (2015), Data Journals: A Survey. J Assn Inf Sci Tec, 66: 1747-1762. doi:10.1002/asi.23358
How does your discipline share data
Does your discipline have a data journal? Or some other mechanism to share data? For example, the American Astronomical Society (AAS) via the publisher IOP Physics offers a supplement series as a way for astronomers to publish data.
Adapted from: Library Carpentry. September 2019. https://librarycarpentry.org/lc-fair-research.
Key Points
Findable, means findable long-term. This requires persistent identifiers (PIDs).
DOIs are one of more common PIDs and can be used to persistently identify software and datasets.
Accessible
Overview
Teaching: 8 min
Exercises: 2 minQuestions
What is a protocol?
What types of protocol are FAIR?
Objectives
Understand what a protocol is
Understand authentication protocols and their role in FAIR
Articulate the value of landing pages
Explain closed, open and mediated access to data
For data & software to be accessible:
A1. (meta)data are retrievable by their identifier using a standardized communications protocol
A1.1 the protocol is open, free, and universally implementable
A1.2 the protocol allows for an authentication and authorization procedure, where necessary
A2. metadata remain accessible, even when the data are no longer available
What is a protocol?
Simply put, it’s an access method of exchanging data over a computer network. Each protocol has its rules for how data is formatted, compressed, checked for errors. Research repositories often use the OAI-PMH or REST API protocols to interface with data in the repository. The following image from TutorialEdge.net: What is a RESTful API by Elliot Forbes provides a useful overview of how RESTful interfaces work:

Hydroshare offers a REST API protocol which will enable many of the functions that are accessible through the web user interface, to be done programmatically:
Wikipedia has a list of commonly used network protocols but check the service you are using for documentation on the protocols it uses and whether it corresponds with the FAIR Principles. For instance, see Hydroshare’s API Instructions page.
Contributor information
Alternatively, for sensitive/protected data, if the protocol cannot guarantee secure access, an e-mail or other contact information of a person/data manager should be provided, via the metadata, with whom access to the data can be discussed. The DataCite metadata schema includes contributor type and name as fields where contact information is included. Collaborative projects such as THOR, FREYA, and ODIN are working towards improving the interoperability and exchange of metadata such as contributor information.
Author disambiguation and authentication
Across the research ecosystem, publishers, repositories, funders, research information systems, have recognized the need to address the problem of author disambiguation. The illustrative example below of the many variations of the name Jens Åge Smærup Sørensen demonstrations the challenge of wrangling the correct name for each individual author or contributor:

Thankfully, a number of research systems are now integrating ORCID into their authentication systems. Zenodo provides the login ORCID authentication option. Once logged in, your ORCID will be assigned to your authored and deposited works.
Exercise to create a Hydroshare Account
- Register for Hydroshare.
- You will receive a confirmation email. Click the link in the email…
- Go to Hydroshare and select Log in.
Understanding whether something is open, free, and universally implementable
Exercise 5
ORCID features a principles page where we can assess where it lies on the spectrum of these criteria. Can you identify statements that speak to these conditions: open, free, and universally implemetable?
Solution
- ORCID is a non-profit that collects fees from its members to sustain its operations Creative Commons CC0 1.0 Universal (CC0) license releases data into the public domain, or otherwise grants permission to use it for any purpose
- It is open to any organization and transcends borders Followup Questions:
- Where can you download the freely available data?
- How does ORCID solicit community input outside of its governance?
- Are the tools used to create, read, update, delete ORCID data open?
Tombstones, a very grave subject
There are a variety of reasons why a placeholder with metadata or tombstone of the removed research object exists including but not limited to staff removal, spam, request from owner, data center does not exist is still, etc. A tombstone page is needed when data and software is no longer accessible. A tombstone page communicates that the record is gone, why it is gone, and in case you really must know, there is a copy of the metadata for the record. A tombstone page should include: DOI, date of deaccession, reason for deaccession, message explaining the data center’s policies, and a message that a copy of the metadata is kept for record keeping purposes as well as checksums of the files.
DataCite offers statistics where the failure to resolve DOIs after a certain number of attempts is reported (see DataCite statistics support pagefor more information). In the case of Zenodo and the GitHub issue above, the hidden field reveals thousands of records that are a result of spam.
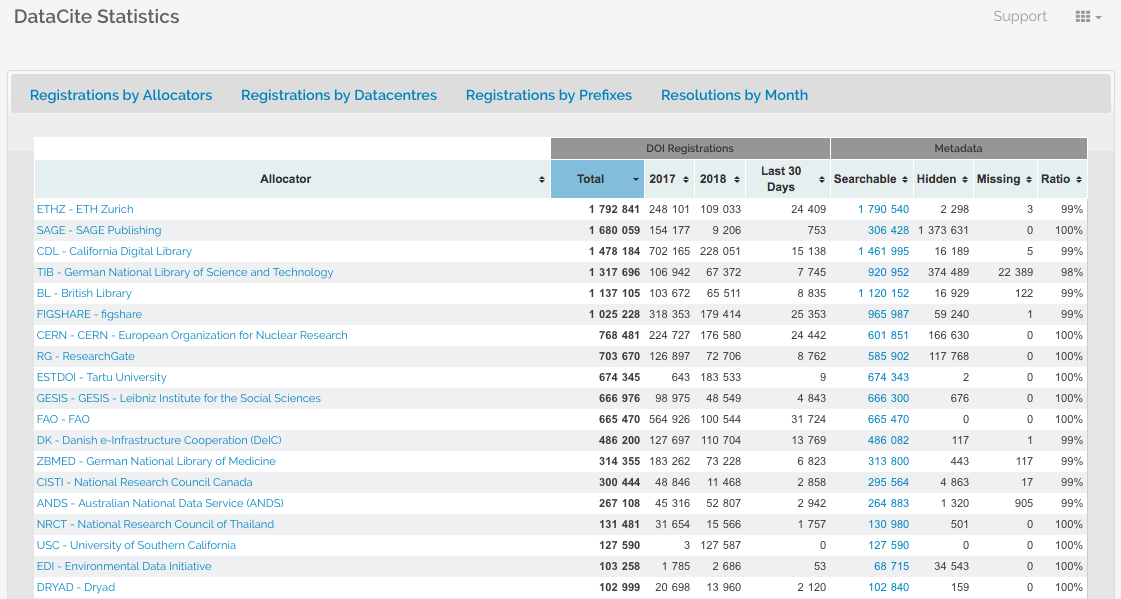
If a DOI is no longer available and the data center does not have the resources to create a tombstone page, DataCite provides a generic tombstone page.
See the following tombstone examples:
- Zenodo tombstone: https://zenodo.org/record/1098445
- Figshare tombstone: https://figshare.com/articles/Climate_Change/1381402
Adapted from: Library Carpentry. September 2019. https://librarycarpentry.org/lc-fair-research.
Key Points
Research repositories often use the OAI-PMH or REST API protocols.
Interoperable
Overview
Teaching: 8 min
Exercises: 0 minQuestions
What does interoperability mean?
What is a controlled vocabulary, a metadata schema and linked data?
How do I describe data so that humans and computers can understand?
Objectives
Explain what makes data and software (more) interoperable for machines
Identify widely used metadata standards for research, including generic and discipline-focussed examples
Explain the role of controlled vocabularies for encoding data and for annotating metadata in enabling interoperability
Understand how linked data standards and conventions for metadata schema documentation relate to interoperability
For data & software to be interoperable:
I1. (meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation
I2. (meta)data use vocabularies that follow FAIR principles
I3. (meta)data include qualified references to other (meta)data
What is interoperability for data and software?
Shared understanding of concepts, for humans as well as machines.
What does it mean to be machine readable vs human readable?
According to the Open Data Handbook:
Human Readable
“Data in a format that can be conveniently read by a human. Some human-readable formats, such as PDF, are not machine-readable as they are not structured data, i.e. the representation of the data on disk does not represent the actual relationships present in the data.”
Machine Readable
“Data in a data format that can be automatically read and processed by a computer, such as CSV, JSON, XML, etc. Machine-readable data must be structured data. Compare human-readable. Non-digital material (for example printed or hand-written documents) is by its non-digital nature not machine-readable. But even digital material need not be machine-readable. For example, consider a PDF document containing tables of data. These are definitely digital but are not machine-readable because a computer would struggle to access the tabular information - even though they are very human readable. The equivalent tables in a format such as a spreadsheet would be machine readable. As another example scans (photographs) of text are not machine-readable (but are human readable!) but the equivalent text in a format such as a simple ASCII text file can machine readable and processable.”
Software uses community accepted standards and platforms, making it possible for users to run the software. Top 10 FAIR things for research software
Describing data and software with shared, controlled vocabularies
See
- https://librarycarpentry.org/Top-10-FAIR//2018/12/01/research-data-management/#thing-8-controlled-vocabulary
- https://librarycarpentry.org/Top-10-FAIR//2019/09/06/astronomy/#thing-6-terminology
- https://librarycarpentry.org/Top-10-FAIR//2018/12/01/historical-research/#thing-6-controlled-vocabularies-and-ontologies
Representing knowledge in data and software
Beyond the PDF
Publishers, librarians, researchers, developers, funders, they have all been working towards a future where we can move beyond the PDF, from ‘static and disparate data and knowledge representations to richly integrated content which grows and changes the more we learn.” Research objects of the future will capture all aspects of scholarship: hypotheses, data, methods, results, presentations etc.) that are semantically enriched, interoperable and easily transmitted and comprehended. Attribution, Evaluation, Archiving, Impact https://sites.google.com/site/beyondthepdf/
Beyond the PDF has now grown into FORCE… Towards a vision where research will move from document- to knowledge-based information flows semantic descriptions of research data & their structures aggregation, development & teaching of subject-specific vocabularies, ontologies & knowledge graphs Paper of the Future https://www.authorea.com/users/23/articles/8762-the-paper-of-the-future to Jupyter Notebooks/Stencilia https://stenci.la/
Making Metadata Interoperable
- provide machine-readable (meta)data with a well-established formalism
- provide as precise & complete metadata as possible
- look for metrics to evaluate the FAIRness of a controlled vocabulary / ontology / thesaurus often do not (yet) exist
- clearly identify relationships between datasets in the metadata (e.g. “is new version of”, “is supplement to”, “relates to”, etc.)
- request support regarding these tasks from the repositories in your field of study for software: follow established code style guides
Examples of Dataste Interoperability:
- Automatic ORCID profile update when DOI is minted – DataCite – CrossRef – ORCID
If others can use your code, convey the meaning of updates with SemVer.org (CC BY 3.0) “version number[ changes] convey meaning about the underlying code” (Tom Preston-Werner)
Linked Data
Top 10 FAIR things: Linked Open Data
Standards: https://fairsharing.org/standards/ schema.org: http://schema.org/
ISA framework: ‘Investigation’ (the project context), ‘Study’ (a unit of research) and ‘Assay’ (analytical measurement) - https://isa-tools.github.io/
Example of schema.org: rOpenSci/codemetar
Modularity http://bioschemas.org
codemeta croswalks to other standards https://codemeta.github.io/crosswalk/
DCAT https://www.w3.org/TR/vocab-dcat/
Using community accepted code style guidelines such as PEP 8 for Python (PEP 8 itself is FAIR)
Scholix - related indentifiers - Zenodo example linking data/software to papers https://dliservice.research-infrastructures.eu/#/ https://authorcarpentry.github.io/dois-citation-data/01-register-doi.html
Key Points
Understand that FAIR is about both humans and machines understanding data.
Interoperability means choosing a data format or knowledge representation language that helps machines to understand the data.
Reusable
Overview
Teaching: 8 min
Exercises: 0 minQuestions
What makes data reusable?
Objectives
Explain machine readability in terms of file naming conventions and providing provenance metadata
Explain how data citation works in practice
Understand key components of a data citation
Explore domain-relevant community standards including metadata standards
Understand how proper licensing is essential for reusability
Know about some of the licenses commonly used for data and software
Exercise 6: Thanks, but no thanks!
In groups discuss:
- Have you ever received data you couldn’t use? Why or why not?
- Have you tried replicating an experiment, yours or someone else? What challenges did you face?
For data & software to be reusable:
R1. (meta)data have a plurality of accurate and relevant attributes
R1.1 (meta)data are released with a clear and accessible data usage licence
R1.2 (meta)data are associated with their provenance
R1.3 (meta)data meet domain-relevant community standards
File naming best practices
A file name should be unique, consistent and descriptive. This allows for increased visibility and discoverability and can be used to easily classify and sort files. Remember, a file name is the primary identifier to the file and its contents.
Do’s and Don’ts of file naming:
Do’s:
- Make use of file naming tools for bulk naming such as Ant Renamer, RenameIT or Rename4Mac.
- Create descriptive, meaningful, easily understood names no less than 12-14 characters.
- Use identifiers to make it easier to classify types of files i.e. Int1 (interview 1)
- Make sure the 3-letter file format extension is present at the end of the name (e.g. .doc, .xls, .mov, .tif)
- If applicable, include versioning within file names
- For dates use the ISO 8601 standard: YYYY-MM-DD and place at the end of the file number UNLESS you need to organise your files chronologically.
- For experimental data files, consider using the project/experiment name and conditions in abbreviations
- Add a README file in your top directory which details your naming convention, directory structure and abbreviations
-
- When combining elements in file name, use common special letter case patterns such as Kebab-case, CamelCase, or Snake_case, preferably use hyphens (-) or underscores (_)
Don’ts:
- When combining elements in file name, use common special letter case patterns such as Kebab-case, CamelCase, or Snake_case, preferably use hyphens (-) or underscores (_)
- Avoid naming files/folders with individual persons names as it impedes handover and data sharing.
- Avoid long names
- Avoid using spaces, dots, commas and special characters (e.g. ~ ! @ # $ % ^ & * ( ) ; < > ? , [ ] { })
- Avoid repetition for ex. Directory name Electron_Microscopy_Images, then you don’t need to name the files ELN_MI_Img_20200101.img
Examples:
- Stanford Libraries guidance on file naming is a great place to start.
- Dryad example:
- 1900-2000_sasquatch_migration_coordinates.csv
- Smith-fMRI-neural-response-to-cupcakes-vs-vegetables.nii.gz
- 2015-SimulationOfTropicalFrogEvolution.R
Directory structures and README files
A clear directory structure will make it easier to locate files and versions and this is particularly important when collaborating with others. Consider a hierarchical file structure starting from broad topics to more specific ones nested inside, restricting the level of folders to 3 or 4 with a limited number of items inside each of them.
The UK data services offers an example of directory structure and naming: https://ukdataservice.ac.uk/manage-data/format/organising.aspx
For others to reuse your research, it is important to include a README file and to organize your files in a logical way. Consider the following file structure examples from Dryad:
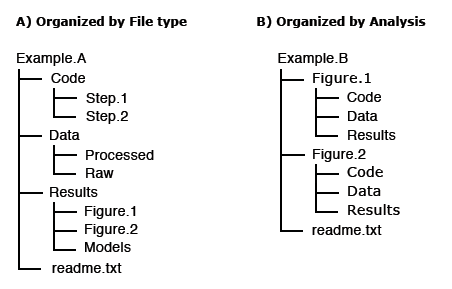
It is also good practice to include README files to describe how the data was collected, processed, and analyzed. In other words, README files help others correctly interpret and reanalyze your data. A README file can include file names/directory structure, glossary/definitions of acronyms/terms, description of the parameters/variables and units of measurement, report precision/accuracy/uncertainty in measurements, standards/calibrations used, environment/experimental conditions, quality assurance/quality control applied, known problems, research date information, description of relationships/dependencies, additional resources/references, methods/software/data used, example records, and other supplemental information.
-
Dryad README file example: https://doi.org/10.5061/dryad.j512f21p
-
Awesome README list (for software): https://github.com/matiassingers/awesome-readme
-
Different Format Types https://data.library.virginia.edu/data-management/plan/format-types/
Disciplinary Data Formats
Many disciplines have developed formal metadata standards that enable re-use of data; however, these standards are not universal and often it requires background knowledge to indentify, contextualize, and interpret the underlying data. Interoperability between disciplines is still a challenge based on the continued use of custom metadata schmes, and the development of new, incompatiable standards. Thankfully, DataCite is providing a common, overarching metadata standard across disciplinary datasets, albeit at a generic vs granular level.
In the meantime, the Research Data Alliance (RDA) Metadata Standards Directory - Working Group developed a collaborative, open directory of metadata standards, applicable to scientific data, to help the research community learn about metadata standards, controlled vocabularies, and the underlying elements across the different disciplines, to potentially help with mapping data elements from different sources.
Metadata Standards Directory
Features: Standards, Extensions, Tools, and Use Cases
Quality Control
Quality control is a fundamental step in research, which ensures the integrity of the data and could affect its use and reuse and is required in order to identify potential problems.
It is therefore essential to outline how data collection will be controlled at various stages (data collection,digitisation or data entry, checking and analysis).
Versioning
In order to keep track of changes made to a file/dataset, versioning can be an efficient way to see who did what and when, in collaborative work this can be very useful.
A version control strategy will allow you to easily detect the most current/final version, organize, manage and record any edits made while working on the document/data, drafting, editing and analysis.
Consider the following practices:
- Outline the master file and identify major files for instance; original, pre-review, 1st revision, 2nd revision, final revision, submitted.
- Outline strategy for archiving and storing: Where to store the minor and major versions, how long will you retain them accordingly.
- Maintain a record of file locations, a good place is in the README files
Example: UK Data service version control guide: https://www.ukdataservice.ac.uk/manage-data/format/versioning.aspx
Research vocabularies
Research Vocabularies Australia https://vocabs.ands.org.au/ AGROVOC & VocBench http://aims.fao.org/vest-registry/vocabularies/agrovoc Dimensions Fields of Research https://dimensions.freshdesk.com/support/solutions/articles/23000012844-what-are-fields-of-research-
Versioning/SHA https://swcarpentry.github.io/git-novice/reference
Binder - executable environment, making your code immediately reproducible by anyone, anywhere. https://blog.jupyter.org/binder-2-0-a-tech-guide-2017-fd40515a3a84
Narrative & Documentation Jupyter Notebooks https://www.contentful.com/blog/2018/06/01/create-interactive-tutorials-jupyter-notebooks/
Licenses From GitHub https://blog.github.com/2015-03-09-open-source-license-usage-on-github-com/
Lack of licenses provide friction, understanding of whether can reuse Peter Murray Project - ContentMine - The Right to Read is the Right to Mine - OpenMinTed Creative Commons Wizard and GitHub software licensing wizards (highlight attribution, non commercial)
Useful content for Licenses Note: TIB Hannover Slides https://docs.google.com/presentation/d/1mSeanQqO0Y2khA8KK48wtQQ_JGYncGexjnspzs7cWLU/edit#slide=id.g3a64c782ff_1_138
Resources
- Choose an open source license: https://choosealicense.com/
- 4 Simple recommendations for Open Source Software https://softdev4research.github.io/4OSS-lesson/
- Top 10 FAIR Imaging https://librarycarpentry.org/Top-10-FAIR//2019/06/27/imaging/
- Licensing your work: https://librarycarpentry.org/Top-10-FAIR//2019/06/27/imaging/#9-licensing-your-work
- The Turing Way a Guide for reproducible Research: https://the-turing-way.netlify.app/welcome
- The Open Science Training Handbook: https://open-science-training-handbook.gitbook.io/book/
- Open Licensing and file formats https://open-science-training-handbook.gitbook.io/book/open-science-basics/open-licensing-and-file-formats#6-open-licensing-and-file-formats
- DCC How to license research data https://www.dcc.ac.uk/guidance/how-guides/license-research-data
Adapted from: Library Carpentry. September 2019. https://librarycarpentry.org/lc-fair-research.
Key Points
It is possible to publish public data that does not meet FAIR standards.
Different fields have variable standards for metadata.
Metadata
Overview
Teaching: 8 min
Exercises: 14 minQuestions
What is metadata?
What do we use metadata for?
Objectives
Recognise what metadata is
Distinguish different types of metadata
Understand what makes metadata interoperable
Know how to decide what to include in metadata
(5 min teaching)
What is (or are) metadata?
Simply put, metadata is data about the data. Sound confusing? Lets clarify: metadata is the description of data. It allows deeper understanding of data and provides insight for its interpretation. Hence, your metadata should be considered as important as your data. Further, metadata plays a very important role in making your data FAIR. It should be continuously added to your research data (not just at the beginning or end of a project!). Metadata can be produced in an automated way (e.g. when you capture a microscopy image usually the accompanying software saves metadata as part of it) or manually.
Let’s take a look at an example:
This is a confocal microscopy image of a C. elegans nematode strain used
as a proteostasis model (Pretty! Isn’t it?). The image is part of the raw data
associated to Goya et al., 2020,
which was deposited in a Public Omero Server
Project
Figure1 set
 Figure credits: María Eugenia Goya
Figure credits: María Eugenia Goya
. What information can you get from the image, without the associated description (metadata)?
Let’s see the associated metadata of the image and the dataset to which it belongs:
Image metadata
Name: OP50 D10Ad_06.czi Image ID: 3485 Owner: Maria Eugenia Goya ORCID: 0000-0002-5031-2470
Acquisition Date: 2018-12-12 17:53:55 Import Date: 2020-04-30 22:38:59 Dimensions (XY): 1344 x 1024 Pixels Type: uint16 Pixels Size (XYZ) (µm): 0.16 x 0.16 x 1.00 Z-sections/Timepoints: 56 x 1 Channels: TL DIC, TagYFP ROI Count: 0
Tags: time course; day 10; adults; food switching; E. coli OP50; NL5901; C. elegans
Dataset metadata
Name: Figure2_Figure2B Dataset ID: 263 Owner: Maria Eugenia Goya ORCID: 0000-0002-5031-2470
Description: The datasets contains a time course of α-syn aggregation in NL5901 C. elegans worms after a food switch at the L4 stage:
E. coli OP50 to OP50 Day 01 adults Day 03 adults Day 05 adults Day 07 adults Day 10 adults Day 13 adults
E. coli OP50 to B. subtilis PXN21 Day 01 adults Day 03 adults Day 05 adults Day 07 adults Day 10 adults Day 13 adults
Images were taken at 6 developmental timepoints (D1Ad, D3Ad, D5Ad, D7Ad, D10Ad, D13Ad)
* Some images contain more than one nematode.
Each image contains ~30 (or more) Z-sections, 1 µmeters apart. The TagYFP channel is used to follow the alpha-synuclein particles. The TL DIC channel is used to image the whole nematode head.
These images were used to construct Figure 2B of the Cell Reports paper (https://doi.org/10.1016/j.celrep.2019.12.078).
Creation date: 2020-04-30 22:16:39
Tags: protein aggregation; time course; E. coli OP50 to B. subtilis PXN21; food switching; E. coli OP50; 10.1016/j.celrep.2019.12.078; NL5901; C. elegans
This is a lot of information!
Types of metadata
According to How to FAIR we can distinguish between three main types of metadata:
- Administrative metadata: data about a project or resource that are relevant for managing it; E.g. project/resource owner, principal investigator, project collaborators, funder, project period, etc. They are usually assigned to the data, before you collect or create them.
- Descriptive or citation metadata: data about a dataset or resource that allow people to discover and identify it; E.g. authors, title, abstract, keywords, persistent identifier, related publications, etc.
- Structural metadata: data about how a dataset or resource came about, but also how it is internally structured. E.g. the unit of analysis, collection method, sampling procedure, sample size, categories, variables, etc. Structural metadata have to be gathered by the researchers according to best practice in their research community and will be published together with the data.
Descriptive and structural metadata should be added continuously throughout the project.
Exercise 6: Identifying metadata types (4 min)
Here we have an excel spreadsheet that contains project metadata for a made-up experiment of plant metabolites
Figure credits: Tomasz Zielinski and Andrés Romanowski
In groups, identify different types of metadata (administrative, descriptive, structural) present in this example.
Solution
- Administrative metadata marked in blue
- Descriptive metadata marked in orange
- Structural metadata marked in green
Figure credits: Tomasz Zielinski and Andrés Romanowski
(6 min teaching)
Where does data end and metadata start?
What is “data” and what is “metadata” can be a matter of perspective: Some researchers’ metadata can be other researchers’ data.
For example, a funding body is categorised as typical administrative metadata, however, it can be used to calculate numbers of public datasets per funder and then used to compare effects of different funders’ policies on open practices.
Adding metadata to your experiments
Good metadata are crucial for assuring re-usability of your outcomes. Adding metadata is also a very time-consuming process if done manually, so collecting metadata should be done incrementally during your experiment.
As we saw metadata can take many forms from as simple as including a ReadMe.txt file, by embedding them inside the Excel files, to using domain specific metadata standards and formats.
But,
- What should be included in metadata?
- What terms should be used in descriptions?
For many assay methods and experiment types, there are defined recommendations and guidelines called Minimal Information Standards.
Minimal Information Standard
The minimum information standard is a set of guidelines for reporting data derived by relevant methods in biosciences. If followed, it ensures that the data can be easily verified, analysed and clearly interpreted by the wider scientific community. Keeping with these recommendations also facilitates the foundation of structuralized databases, public repositories and development of data analysis tools. Individual minimum information standards are brought by the communities of cross-disciplinary specialists focused on issues of the specific method used in experimental biology.
Minimum Information for Biological and Biomedical Investigations (MIBBI) is the collection of the most known standards.
FAIRSharing offers excellent search service for finding standards
Exercise 7: Minimal information standard example (2 min)
Look at Minimum Information about a Neuroscience Investigation (MINI) Electrophysiology Gibson, F. et al. Nat Prec (2008). which contains recommendations for reporting the use of electrophysiology in a neuroscience study.
(Neuroscience (or neurobiology) is the scientific study of the nervous system).Scroll to Reporting requirement and decide which of the points 1-8 are:
- a) important for understanding and reuse of data
- b) important for technical replication
- c) could be applied to other experiments in neuroscience
Solution
Possible answers:
- a) 2, 3, 4, 5, 6, 8a-b
- b) 3, 7
- c) 2, 3, 4, 5, 6
What if there are no metadata standards defined for your data / field of research?
Think about the minimum information that someone else (from your lab or from any other lab in the world) would need to know to be able to work with your dataset without any further input from you.
Think as a consumer of your data not the producer!
Exercise 8: What to include - discussion (2 minutes)
Think of the data you generate in your projects, and imagine you are going to share them.
What information would another researcher need to understand or reproduce your data (the structural metadata)?
For example, we believe that any dataset should have:
- a name/title
- its purpose or experimental hypothesis
Write down and compare your proposals, can we find some common elements?
Solution
Some typical elements are:
- biological material, e.g. Species, Genotypes, Tissue type, Age, Health conditions
- biological context, e.g. speciment growth, entrainment, samples preparation
- experimental factors and conditions, e.g. drug treatments, stress factors
- primers, plasmid sequences, cell line information, plasmid construction
- specifics of data acquisition
- specifics of data processing and analysis
- definition of variables
- accompanying code, software used (version nr), parameters applied, statistical tests used, seed for randomisation
- LOT numbers
Metadata and FAIR guidelines
Metadata provides extremely valuable information for us and others to be able to interpret, process, reuse and reproduce the research data it accompanies.
Because metadata are data about data, all of the FAIR principles i.e. Findable, Accessible, Interoperable and Reusable apply to metadata.
Ideally, metadata should not only be machine-readable, but also interoperable so that they can interlink or be reasoned about by computer systems.
Attribution
Content of this episode was adapted from:
Key Points
Metadata provides contextual information so that other people can understand the data.
Metadata is key for data reuse and complying with FAIR guidelines.
Metadata should be added incrementally through out the project
Public repositories
Overview
Teaching: 8 min
Exercises: 2 minQuestions
Where can I deposit datasets?
What are general data repositories?
How to find a repository?
Objectives
See the benefits of using research data repositories.
Differentiate between general and specific repositories.
Find a suitable repository.
What are research data repositories?
(13 min teaching) Research data repositories are online repositories that enable the preservation, curation and publication of research ‘products’. These repositories are mainly used to deposit research ‘data’. However, the scope of the repositories is broader as we can also deposit/publish ‘code’ or ‘protocols’ (as we saw with protocols.io).
There are general “data agnostic” repositories, for example:
Or domain specific, for example:
- UniProt protein data,
- GenBank sequence data,
- MetaboLights metabolomics data
- GitHub for code.
- Hydroshare for water data.
Research outputs should be submitted to discipline/domain-specific repositories whenever it is possible. When such a resource does not exist, data should be submitted to a ‘general’ repository. Research data repositories are a key resource to help in data FAIRification as they assure Findability and Accessibility.
Exercise 9: Public general record (8 min)
Have a look at the following record for data set in Hydroshare repository: Hydroshare. What elements make it FAIR?
Solution
The elements that make this deposit FAIR are:
Findable (persistent identifiers, easy to find data and metadata):
- F1. (Meta)data are assigned a globally unique and persistent identifier - YES
- F2. Data are described with rich metadata (defined by R1 below)- YES
- F3. Metadata clearly and explicitly include the identifier of the data they describe - YES
- F4. (Meta)data are registered or indexed in a searchable resource - YES
Accessible (The (meta)data retrievable by their identifier using a standard web protocols):
- A1. (Meta)data are retrievable by their identifier using a standardised communications protocol - YES
- A2. Metadata are accessible, even when the data are no longer available - YES
Interoperable (The format of the data should be open and interpretable for various tools):
- I1. (Meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation. - YES
- I2. (Meta)data use vocabularies that follow FAIR principles - PARTIALLY
- I3. (Meta)data include qualified references to other (meta)data - YES
Reusable (data should be well-described so that they can be replicated and/or combined in different settings, reuse states with a clear licence):
- R1. (Meta)data are richly described with a plurality of accurate and relevant attributes - YES
- R1.1. (Meta)data are released with a clear and accessible data usage license - YES
- R1.2. (Meta)data are associated with detailed provenance - YES
- R1.3. (Meta)data meet domain-relevant community standards - YES/PARTIALLY
(4 min discussion)
Minimal data set
Minimal data set to consist of the data required to replicate all study findings reported in the article, as well as related metadata and methods.
- The values behind the means, standard deviations and other measures reported;
- The values used to build graphs;
- The points extracted from images for analysis.
(no need for raw data if the standard in the field is to share data that have been processed)
How do we choose a research data repository?
(3 min teaching) As a general rule, your research needs to be deposited in discipline/data specific repository. If no specific repository can be found, then you can use a generalist repository. Having said this, there are tons of data repositories to choose from. Choosing one can be time consuming and challenging as well. So how do you go about finding a repository:
- Check the publisher’s / funder’ recommended list of repositories, some of which can be found below:
- Check Fairsharing recommendations
- alternatively, check the Registry of research data repositories - re3data
Exercise 10: Finding a repository (5 min + 4 min discussion).
a) Find a repo for genomics data.
b) Find a repo for microscopy data.
Note to instructor: Fairsharing gives few options, people may give different answer follow up why they selected particular ones.Solution
a) GEO/SRA and ENA/ArrayExpress are good examples. Interestingly these repositories do not issue a DOI.
b) IDR is good examples.
(6 min teaching)
A list of UoE BioRDM’s recommended data repositories can be found here.
What comes first? the repository or the metadata?
Finding a repository first may help in deciding what metadata to collect and how!
Extra features
It is also worth considering that some repositories offer extra features, such as running simulations or providing visualisation. For example, FAIRDOMhub can run model simulations and has project structures. Do not forget to take this into account when choosing your repository. Extra features might come in handy.
Can GitHub be cited?
To make your code repositories easier to reference in academic literature, you can create persistent identifiers for them. Particularly, you can use the data archiving tool in Zenodo to archive a GitHub repository and issue a DOI for it.
Evaluating a research data repository
You can evaluate the repositories by following this criteria:
- who is behind it, what is its funding
- quality of interaction: is the interaction for purposes of data deposit or reuse efficient, effective and satisfactory for you?
- take-up and impact: what can I put in it? Is anyone else using it? Will others be able to find stuff deposited in it? Is the repository linked to other data repositories so I don’t have to search tehre as well? Can anyone reuse the data? Can others cite the data, and will depositing boost citations to related papers?
- policy and process: does it help you meet community standards of good practice and comply with policies stipulating data deposit?
Resources
- An interesting take can be found at Peter Murray-Rust’s blog post Criteria for succesful repositories.
Attribution
Content of this episode was adapted or inspired by:.
Key Points
Repositories are the main means for sharing research data.
You should use data-type specific repository whenever possible.
Repositories are the key players in data reuse.
Exercises
Overview
Teaching: 0 min
Exercises: 10 minQuestions
What makes this dataset FAIR?
Objectives
Analyze a dataset to see if it is FAIR.
(10 min exercise)
Exercise 11: What aspect of this dataset are FAIR? (10 minutes)
At bare minimum, any dataset can probably benefit from having the below information available:
- a name/title
- its purpose or experimental hypothesis
Analyze the below dataset from HydroShare. Hydroshare: Annual soil moisture predictions across conterminous United States using remote sensing and terrain analysis across 1 km grids (1991-2016)
Use the ARDC FAIR self assessment tool
Solution
Solutions will probably contain the following:
- Findable: mostly FAIR
- Accessible: mostly FAIR
- Interoperable: mostly FAIR
- Reusable: mostly FAIR
Exercise 12: What aspect of this dataset are FAIR? (10 minutes)
Analyze the below dataset from HydroShare. Long-term, gridded standardized precipitation index for Hawai‘i
Use the ARDC FAIR self assessment tool
Solution
Solutions will probably contain the following:
- Findable: mostly FAIR
- Accessible: mostly FAIR
- Interoperable: mostly FAIR
- Reusable: mostly FAIR
Exercise 13: What aspect of this dataset are FAIR? (10 minutes)
Analyze the below dataset from HydroShare. ‘Ike Wai: Groundwater Chemistry - Nutrient Data
Use the ARDC FAIR self assessment tool
Solution
Solutions will probably contain the following:
- Findable: mostly FAIR
- Accessible: mostly FAIR
- Interoperable: mostly FAIR
- Reusable: mostly FAIR
Attribution
Content of this episode was adapted from:
Key Points
A spectrum exists for FAIR data sharing.
Ethics
Overview
Teaching: 8 min
Exercises: 0 minQuestions
What ethical considerations are there when making data public?
Objectives
Understand privacy, freedom, explainability, and fairness as they go into managing data ethics.
Technology poses ethical challenges. There are several areas of research in data ethics. This will serve as a primer for them, additional reading is included.
- Privacy
- Freedom
- Explainability
- Fairness
Privacy what control do people have over the data collected about them? For example, if a person has a BRCA mutation that increases their risk of cancer should their health insurer be able to increase the insurance rate of their policy?
Freedom do people have the freedom to share data about themselves without fear of consequences?
Explainability can the underlying algorithmic processes be explained or tested?
Are you or do you know anyone who believes their phone is listening to them?
- Companies who implement predictive analytics are drifting towards needing to prove the negative with regard to privacy (that they’re following their privacy policies) to consumers because their predictive analytics are so good.
Fairness Because machine learning algorithms often find new, unexpected connections. Such algorithms are useful because they can interpret data that a human cannot; they can improve the fairness of these decisions or they can exacerbate existing biases. Ascertaining when and how machine learning systems introduce bias into decision-making processes presents a significant new challenge in developing these tools.
“While we don’t promise equal outcomes, we have strived to deliver equal opportunity.” –Barack Obama
- Humans do not entirely agree on what is fair.
Choice of Algorithmic Model Impacts the Output. Low Impact Decision, High Volume ex. Facebook Advertising High Impact Decision, Low Volume ex. Medical Testing
Anti-discrimination laws cover machine learning algorithms and, even if variables related to protected-class status are excluded, these algorithms can still produce disparate impacts. Such impacts can be measured if the variables that they use correlate with both the output variable and a variable for protected-class status. Even unintentional discrimination results in legal risk.
Both public and private entities should conduct disparate impact assessments. Software developers should perform disparate impact analyses before publishing or using their algorithms.
Resources
- https://www.brookings.edu/research/fairness-in-algorithmic-decision-making/
Key Points
Privacy, freedom, explainability, and fairness all go into managing data ethics.
Security
Overview
Teaching: 8 min
Exercises: 0 minQuestions
What measures will you take to secure your data?
Objectives
Discuss steps and changes in your habits you will take after learning about data security.
Today, we will approach two different aspcets of data security.
- Securing data from hostile people and groups that would compromise the system.
- Securing data from general threats such as power outages, fires, floods, and hardware failure.
Securing data from hostile groups: Types of attacks
-
Malware: Malicious software. Malware is activated when a user clicks on a malicious link or attachment, which leads to installing dangerous software. Cisco reports that malware, once activated, can: -Block access to key network components (ransomware) -Install additional harmful software -Covertly obtain information by transmitting data from the hard drive (spyware) -Disrupt individual parts, making the system inoperable
-
Emotet: The Cybersecurity and Infrastructure Security Agency (CISA) describes Emotet as “an advanced, modular banking Trojan that primarily functions as a downloader or dropper of other banking Trojans. Emotet continues to be among the most costly and destructive malware.”
-
Denial of Service: A denial of service (DoS) is a type of cyber attack that floods a computer or network so it can’t respond to requests. A distributed DoS (DDoS) does the same thing, but the attack originates from a computer network. Cyber attackers often use a flood attack to disrupt the “handshake” process and carry out a DoS. Several other techniques may be used, and some cyber attackers use the time that a network is disabled to launch other attacks. A botnet is a type of DDoS in which millions of systems can be infected with malware and controlled by a hacker, according to Jeff Melnick of Netwrix, an information technology security software company. Botnets, sometimes called zombie systems, target and overwhelm a target’s processing capabilities. Botnets are in different geographic locations and hard to trace.
-
Man in the Middle: A man-in-the-middle (MITM) attack occurs when hackers insert themselves into a two-party transaction. After interrupting the traffic, they can filter and steal data, according to Cisco. MITM attacks often occur when a visitor uses an unsecured public Wi-Fi network. Attackers insert themselves between the visitor and the network, and then use malware to install software and use data maliciously.
-
Phishing: Phishing attacks use fake communication, such as an email, to trick the receiver into opening it and carrying out the instructions inside, such as providing a credit card number. “The goal is to steal sensitive data like credit card and login information or to install malware on the victim’s machine,” Cisco reports.
-
SQL Injection: A Structured Query Language (SQL) injection is a type of cyber attack that results from inserting malicious code into a server that uses SQL. When infected, the server releases information. Submitting the malicious code can be as simple as entering it into a vulnerable website search box.
-
Password Attacks: With the right password, a cyber attacker has access to a wealth of information. Social engineering is a type of password attack that Data Insider defines as “a strategy cyber attackers use that relies heavily on human interaction and often involves tricking people into breaking standard security practices.” Other types of password attacks include accessing a password database or outright guessing.
An Arms Race
Cyber security practices continue to evolve as the internet and digitally dependent operations develop and change. Data storage on devices such as laptops and cellphones makes it easier for cyber attackers to find an entry point into a network through a personal device. For example, in the May 2019 book Exploding Data: Reclaiming Our Cyber Security in the Digital Age, former U.S. Secretary of Homeland Security Michael Chertoff warns of a pervasive exposure of individuals’ personal information, which has become increasingly vulnerable to cyber attacks. Increase in both data use and data sharing are contributing to cybersecurity threats. Volume and connectedness both create new areas to exploit.
Strategies to Prevent Hostile Groups from Stealing Data:
- Complex Passwords
- Setting 2 Factor Authentication
- Choosing a Reputible Data Repository
Securing Data from Other Natural Events Data loss can happen for many reasons such as fires, floods, and hardware failure. The main strategy for preventing this is data redundancy, where data is stored in multiple locations.
Large datasets, will often span multiple hard disks as we approach 1 terabit per square inch (though to be the superparamagnetic limit) of storage density in magnetic hard drives with increasing data size. Storage arrays are ften redundent, redundancy is often created using various types of RAID configurations.
- Raid 1 is an exact copy (or mirror) of a set of data on two or more disks.
- Raid 5 consists of block-level striping with distributed parity and requires that all drives but one be present to operate. Upon failure of a single drive, subsequent reads can be calculated from the distributed parity such that no data is lost. RAID 5 requires at least three disks.
Small datasets and files are often stored on laptops and computer hard drives. While storage on these devices is more reliable than ever, the data could be lost in the event of damage to the computer. Data storage is affordable, recreating data is not! Important files should never be in only 1 place. Cloud storage can be useful for this. Some cloud storage services:
- Dropbox
- GSuite
- OneDrive
- iCloud
Resources
Attribution RAID The BioRDM team has a lot of information about the here taught course material on their BioRDM wiki.
Key Points
There are simple steps to help make your data more secure
Implement these steps to avoid data loss